In cells, a high concentration of actin monomers is “sequestered” by binding proteins, preventing the assembly of filamentous structures. At the same time, monomers that can polymerize work with binding proteins formin and Arp 2/3 to assemble linear and branched networks, respectively, as shown in the diagram below.
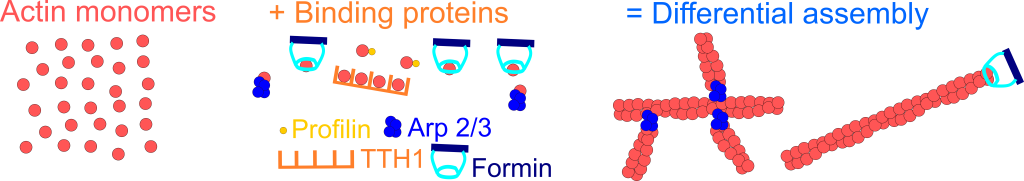
Recent work showed that a limiting actin pool might contribute to disassembly, as cells that lacked functional Arp 2/3 complex assembled more linear actin structures (see below video).
Our main line of work is to implement stochastic (Gillespie) simulations to understand what conditions favor branched vs. linear assembly from a limiting pool of actin monomers. This work is in collaboration with members of the Kovar lab at University of Chicago, who are performing in vitro experiments to test our model predictions.