To simulate the cell cytoskeleton, we use a coarse-grained microscopic description, where each actin filament is represented explicitly inside of the surrounding viscous fluid. From a computational perspective, the main challenges for simulating actin filaments are
- Multiple length scales (actin is about 1000 times longer than it is wide)
- Constraints on the dynamics (actin filaments are inextensible)
- Hydrodynamic interactions (filaments moving in a common fluid generate flows which move the other filaments)
- Brownian motion (filaments are semiflexible, so their bending fluctuations have to be simulated in a way that satisfies the theorems of equilibrium statistical mechanics).
For linear actin filaments, we addressed these challenges in prior work. Our present focus is to extend our work to branched fiber architectures, which have nonlocal constraints (meaning the constraints between two or more filaments are coupled together through the branch point, see below). This work also applies to strongly cross linked filaments.
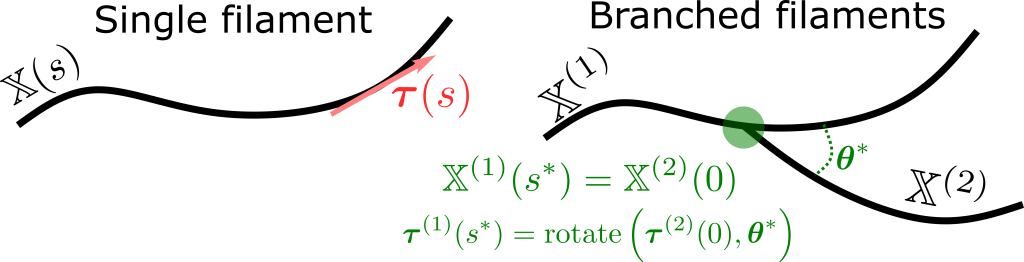
By modeling branches and cross linkers as constraints instead of linear springs, we are able to take longer time steps and simulate larger systems for longer. However, each time step is more expensive, since we have to solve larger linear systems when fibers are coupled together. We are exploring preconditioning strategies to accelerate these linear solves, ultimately making our algorithms adaptable to the kinds of cytoskeletal architectures encountered in live cells.