*Note to all users: As of 07/14/2019, all posts have been moved to (and are being updated at crivaldi.github.io . I don’t have plans to shut this site down, but I have no idea how long it will be maintained by the university in the future.*
String manipulation with sed and grep
What is string manipulation and why do we care about this?
As biologists, or more generally as people interested in working with data, a lot of what we want to do will require us to manipulate many characters at the same time.
By definition:
- A character is class whose instances can hold a single character value.
- A string is an immutable class for working with multiple characters.
For our purposes, we can consider strings as information that we don’t want to use for numerical calculations. DNA sequences, column or row names, and categorical/qualitative data values will generally be strings. You might want to remove the primers from a lot (like a lot) of sequences at the same time, or you might want to remove whitespaces from a dataset you found online.
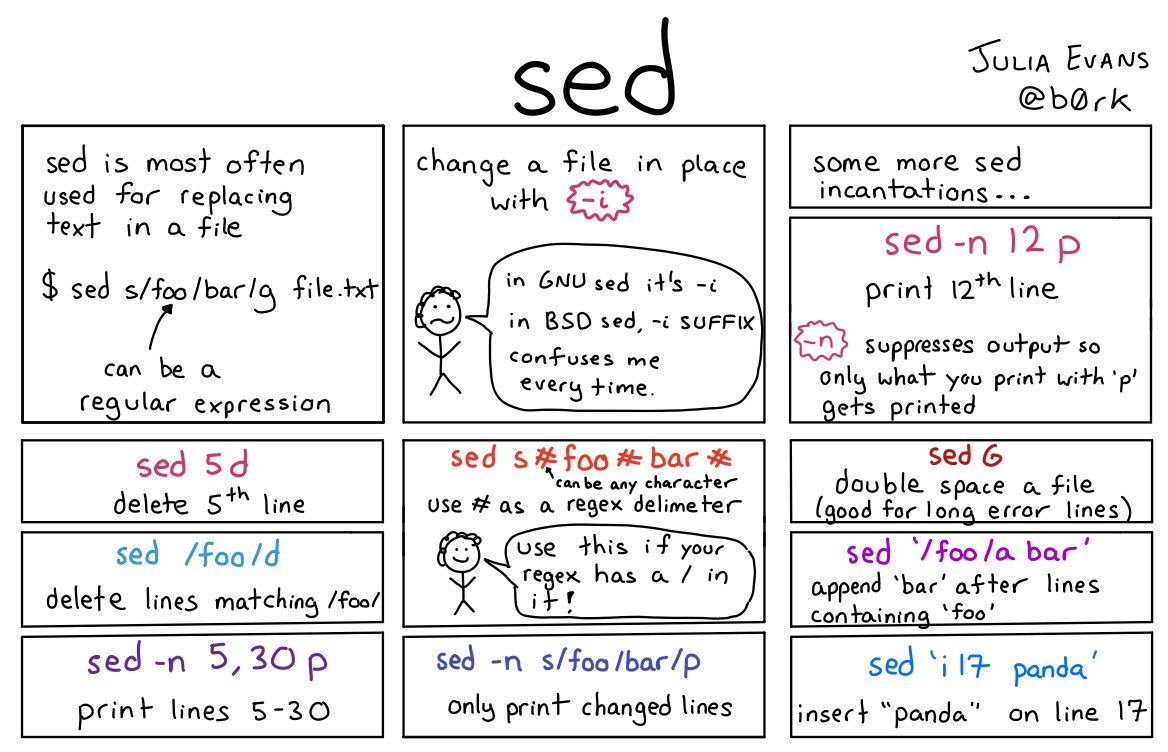
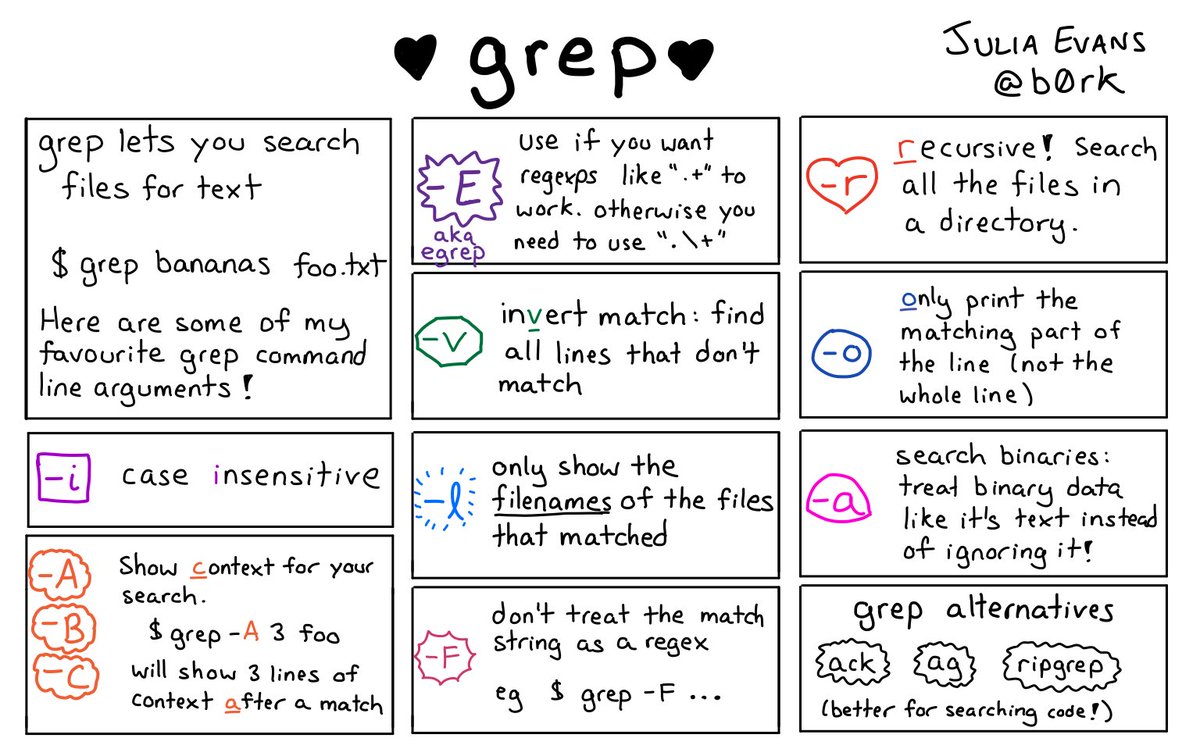
Most of our string manipulation is covered by the previous links that are tied in with the for loops – here are a couple of useful comics for some of those commands though. ‘awk’ is new but you’re probably going to run into it during google adventures. As it in some ways is its own programming language, it’s very much worth learning, we just didn’t quite have time to fit it into our classwork.

https://pythonforbiologists.com/printing-and-manipulating-text/