The Multicellular Systems Lab’s Github Page is here.
NEW! MAPPER: An open-source, high-dimensional image analysis pipeline is available on Github.
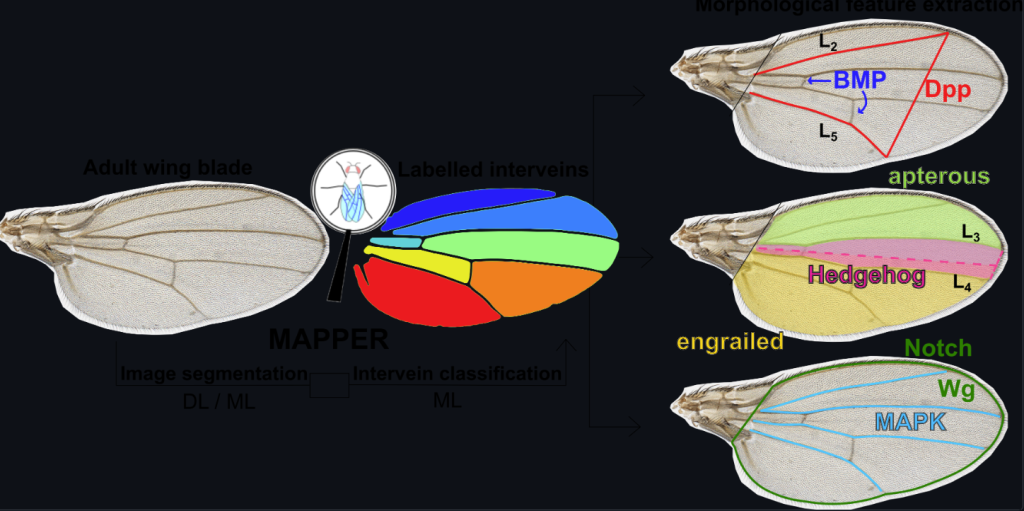
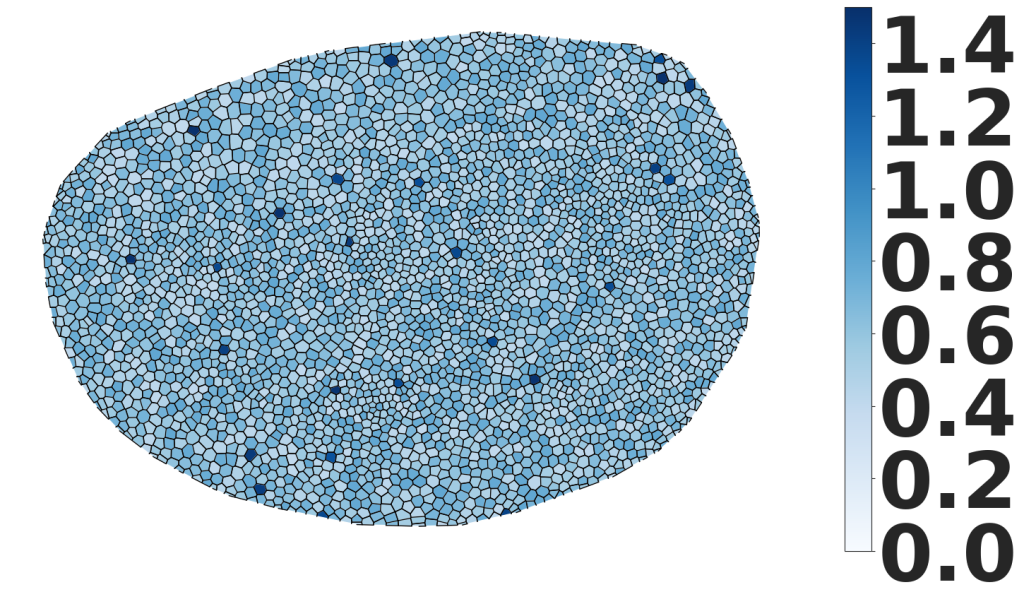
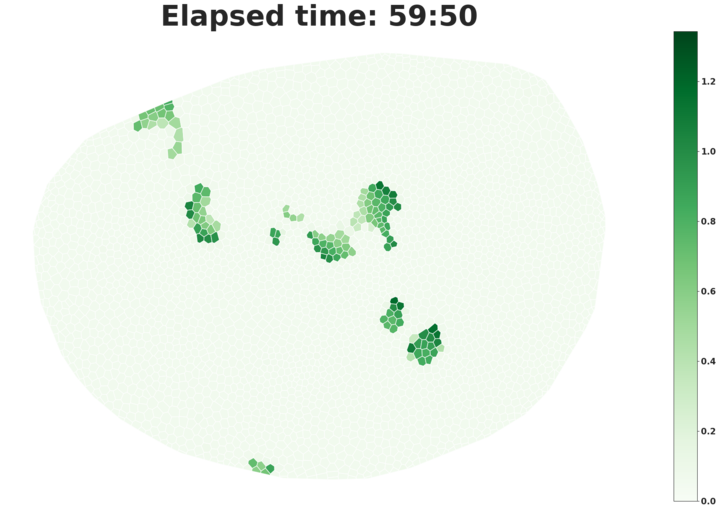
We also have a demo of our calcium signaling project available through Colab here.
Affiliations:

We are part of the new NSF-funded Biology Integration Institute Emergent Mechanisms in Biology of Robustness Integration and Organization Institute (EMBRIO)!
The Zartman lab is affilliated with the Mike and Josie Harper Cancer Research Institute , Notre Dame’s Advanced Diagnostics and Therapeutics (AD&T) initiative, and Notre Dame’s Center for Stem Cells and Regenerative Medicine.
Research links:
Google Scholar Page: Zartman, J.J.
CalciumInsights GitHub Repository
CalciumInsights is an open-source R/Shiny application developed by the EMBRIO Institute for analyzing calcium imaging data. The tool is designed to be tissue-agnostic and provides interactive visualization and statistical analysis of calcium traces. The repository includes source code, documentation, and installation instructions, enabling researchers to run and adapt the app for their own datasets.
The EMBRIO Data Inventory on PURR (Purdue University Research Repository) is a centralized digital catalog of datasets generated by the EMBRIO Institute, including contributions from our lab. It provides access to raw and processed files, detailed metadata, version histories, and usage licenses. Researchers can use the inventory to identify datasets, explore experimental context such as design and sample types, and trace data provenance. By making our lab’s data and EMBRIO-wide research outputs findable, accessible, interoperable, and reusable (FAIR), this resource enhances transparency, reproducibility, and collaboration.
The EMBRIO Modeling Course is a comprehensive documentation resource covering computational modeling methods relevant to developmental biology. It includes tutorials, examples, and explanatory materials drawn from the EMBRIO Institute’s modeling efforts. The repository (on GitHub) supports those interested in building, running, or understanding models of biological systems — especially embryo modeling. It aims to guide researchers through workflows, best practices, and technical details, making modeling tools more accessible and easier to integrate into lab projects.
Links to previous workshops and conferences that we have (co-)organized:
2025 Joint Q-Bio symposium at Notre Dame
