Author: Megan Vahsen
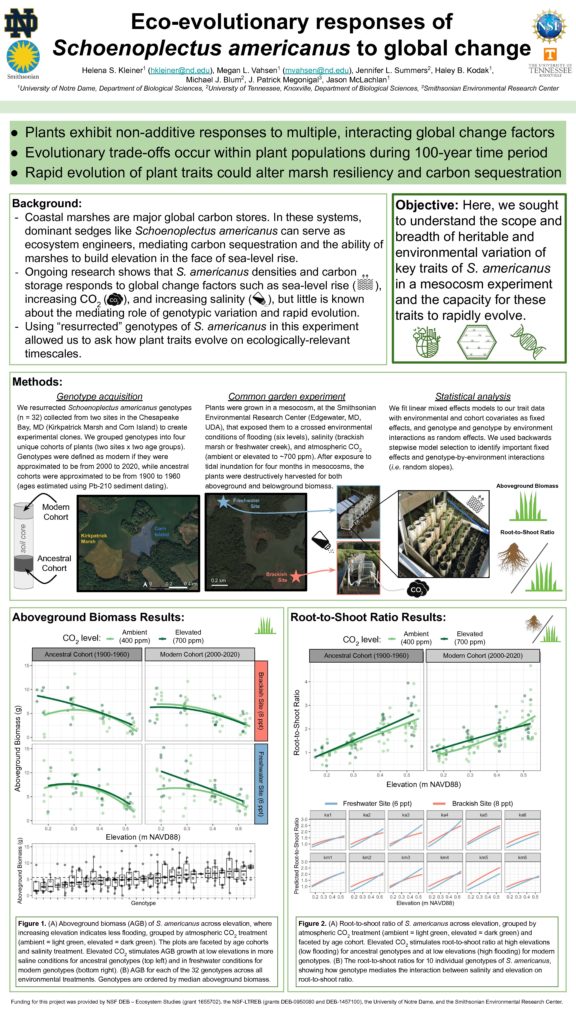
In early October, several members of the McLachlan lab conducted a harvest of our mesocosm experiment at the Smithsonian Environmental Research Center (SERC) in Edgewater, MD. The experiment was a huge effort as a part of our ‘Blue Genes’ NSF grant (in collaboration with the Blum lab at the University of Tennessee Knoxville and the Megonigal lab at SERC) to investigate how intraspecific variation and evolution contribute to ecosystem-level processes. Previous experiments from our lab suggest that there is genetically-based variation in functional traits of a coastal marsh sedge (Schoenoplectus americanus) important to marsh accretion. Here, we investigated how genotype by environment interactions influenced phenotypic variation in a variety of functional traits. We exposed clones of 36 resurrected genotypes of S. americanus to a variety of environmental factors: elevated CO2, salinity, flooding, and interspecific competition. Resurrected genotypes were collected from varying depths of sediment cores across locations in the Chesapeake Bay.
We grew S. americanus genotype clones in PVC pots placed in ‘marsh organs’ (i.e. racks with positions that vary by elevation, Fig 1). We enclosed organs with open-air chambers which allowed us to manipulate atmospheric CO2. This experimental set-up was replicated at two sites: one at the Global Change Research Wetland (higher salinity) and one in a nearby creek (lower salinity, Fig 2). For some pots, S. americanus grew in the presence of an interspecific competitor, Spartina patens, a high marsh, less flood-tolerant species. In June 2019, we established plants in 670 PVC pots across 14 organs.
The harvest in October was a long week of hard work that resulted in a bounty of data to be analyzed! Megan Vahsen, Haley Kodak, and Jason McLachlan from the McLachlan lab, Jenn Summers from UTK, and Helena Kleiner from SERC processed thousands of stems (Figs 3 &4). We were lucky to have over 10 volunteers outside of our team that helped with measuring and recording— it was a real team effort! For each PVC pot, we measured heights and widths of each stem, harvested aboveground biomass, and stored PVC pots in a freezer for processing of belowground biomass which will occur this fall and winter. Jenn and Haley also collected samples for RNA and epigenetic analysis, at harvest. This harvest data is just some of the interesting data we collected for this experiment. Over four field campaigns, Helena (SERC) took porometry measurements on a subset of plants to capture stomatal conductance. Tom Mozdzer (Bryn Mawr) took greenhouse gas flux measurements on a subset of pots and sampled soil for later microbial analysis. Finally, we took porewater samples from a subset of pots for nitrate concentration analysis that Helena is leading (Fig 5). From this experiment, we hope to have a broad picture of how intraspecific variation, evolution, and genotype by environment interactions influence ecosystem processes.

Figure 1. View of experimental plants from the top of an open-air chamber. Pots are placed at different elevations to allow for different levels of flooding as tides move in and out of the creek. Photo credit: Helena Kleiner.

Figure 2. Drone-captured view of the newly built freshwater site. CO2 blowers and sensors were all run on solar power. Helena Kleiner (SERC) led the construction of the site. Photo credit: David Klinges.

Figure 3. Haley, Megan and Jenn measuring plants at the GCREW (high salinity) site. A small unicorn piñata was a good luck charm for the harvest. Photo credit: Helena Kleiner.

Figure 4. Jason was a huge help shuffling pots back and forth from the marsh organs to processing tables. Photo credit: Helena Kleiner.

Figure 5. Porewater sippers in PVC pots.